Publication list (supported by NSF-PGRP awards: 2002-2020)
Biological Resources Available from Research Supported by NSF Plant Genome Research Program Awards IOS-0077622, IOS-0605059, IOS-1025642, and IOS-1546625
All resources in the publications supported by the NSF Plant Genome Research Program (listed below) are available upon request.
Resources include:
Contact for biological materials: Wei Zhang
Videos are available for the PTI assays and VIGS assays associated with Chakravarthy et al. (2009) and Velásquez et al. (2009), respectively.
Seeds of many tomato accessions are available from the Tomato Genetics Resource Center (TGRC) at the University of California-Davis. A collection of Solanum lycopersicum (M82) x S. pennellii (LA716) introgression lines, which are useful for mapping natural variation in many traits including plant immunity, are described and available for ordering: (TGRC - S. pennellii Introgression Lines)
Tomato RNAseq data
Pombo, M.A., et al. 2014. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome Biology 15:492
- Table S1. ETI-, PTI- and common-induced gene expression data (Excel)
- Table S2. GO term analysis of ETI-, PTI- and common induced genes (Excel)
Rosli, H. G., et al 2013. Transcriptomics-based screen for genes induced by flagellin and repressed by pathogen effectors identifies a cell wall-associated kinase involved in plant immunity. Genome Biol. 14:R149.
contact for biological materials: Greg Martin
MAMP response among tomato cultivars
Veluchamy, S., et al. 2014. Natural variation for responsiveness to flg22, flgII-28, and csp22 and Pseudomonas syringae pv. tomato in heirloom tomatoes. PloS one 9:e106119.
Table S4. Summary of responses to MAMPs and disease in the field (Doc)
Tomato Kinome data
Singh, D. K., et al. 2013. The tomato kinome and the TOKN ORFeome: novel resources for the study of kinases and signal transduction in tomato and Solanaceae. Mol Plant Microbe Interact.
- Predicted tomato kinome (supp Table 1) (Excel)
- TOKN 1.0 tomato kinase library (supp Table 3) (Excel)
- TOKN 1.0 kinases in tomato protein microarrray (supp Table 4) (Excel)
contact for biological materials: Sorina Popescu
Transcription Factor Binding Site Prediction Method
Saha, S., and Lindeberg, M. 2013. Bound to Succeed: Transcription Factor Binding-site Prediction and Its Contribution to Understanding Virulence and Environmental Adaptation in Bacterial Plant Pathogens. Mol Plant Microbe Interact 26:1123-1130.
This review of transcription factor binding sites characterized in phytopathogenic bacteria includes description of Pred_cutoff, a computational method for quantitative statistical comparison of the performance of different tools for predicting TFBSs. Pred_cutoff scripts and alignments for 14 TFBS in phytopathogenic bacteria are available at Github (here)
Nicotiana benthamiana Draft Genome Sequence
Bombarely, A., Rosli, H. G., Vrebalov, J., Moffett, P., Mueller, L. A., and Martin, G. B. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact 12:1523-30
Nicotiana benthamiana genome project: http://bti.cornell.edu/nicotiana-benthamiana/
Plant Defense Genes: Nicotiana benthamiana
Chakravarthy et al, 2010. Identification of Nicotiana benthamiana genes involved in PAMP-triggered immunity (PMID:20459311)
Genes comprising the immunity-induced gene collection identified in tomato (Excel)
Nicotiana benthamiana genes implicated in PTI - BLAST analysis (Excel)
Model summarizing the possible roles of genes linked to PAMP-triggered immunity
(click to see larger image)
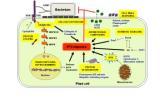
Nicotiana benthamiana genes compromising PTI when silenced
N. benth
gene silenced |
Category |
source |
Percentage of plants showing compromised PTI with the indicated inducer/challenger combination |
Pf/DC |
Pp/DC |
Pf/Q1-1 |
Agro/Ptab |
TRV2 |
- |
- |
0% |
0% |
0% |
0% |
BAK1 |
signaling |
VIGS library |
53% |
22% |
44% |
3% |
FLS2 |
PRR |
VIGS library |
75% |
48% |
66% |
5% |
ALDH |
secondary metabolism |
VIGS library |
8% |
4% |
21% |
8% |
Cathepsin B |
protease |
VIGS library |
17% |
17% |
33% |
8% |
Cyclophilin |
protein folding |
VIGS library |
33% |
26% |
44% |
4% |
Cytochrome C |
secondary metabolism |
VIGS library |
32% |
8% |
32% |
4% |
Drm-3 |
hormone signalling |
VIGS library |
45% |
11% |
17% |
6% |
HCBT |
secondary metabolism |
VIGS library |
20% |
24% |
16% |
8% |
Plastocyanin |
secondary metabolism |
VIGS library |
29% |
13% |
38% |
13% |
Alternative oxidase |
energy metabolism |
immunity induced |
20% |
13% |
40% |
7% |
Anionic peroxidase |
cell wall protein |
|
42% |
47% |
47% |
5% |
CA4H |
secondary metabolism |
|
43% |
21% |
50% |
29% |
EDS1 |
defense signaling |
|
27% |
40% |
27% |
7% |
proteasome 26S subunit |
protein stability |
|
36% |
27% |
0% |
9% |
Trunsducin |
signaling |
|
8% |
8% |
15% |
46% |
Ubiquitin activatinng enzyme |
protein stability |
|
12% |
18% |
29% |
25% |
Percentages in bold represent those combinations where PTI was compromised as determined by pairwise comparisons of the TRV2 control with each of the candidate genes with a Fisher’sexact test (α = 0.05).
Abbreviations: HCBT = Anthranilate N hydroxycinnamoyl/benzoyltransferase, ALDH = Aldehyde dehydrogenase, Agro = A.
tumefaciens, DC = Pst DC3000, Pf = P. fluorescens, Pp = P. putida, Ptab = P. s. pv. tabaci, Q1-1 = Pst DC3000 deltahopQ1-1.
Extent of cell death in Nicotiana benthamiana caused by inoculation with different challengers
N. benth
gene silenced |
Extent of cell death upon
inoculation with Pto DC3000 |
Extent of cell death upon
inoculation with Pto DC3000 deltahopQ1-1 |
Full |
Partial |
None |
Full |
Partial |
None |
TRV2 |
8 |
3 |
1 |
2 |
1 |
5 |
BAK1 |
1 |
3 |
4 |
4 |
4 |
0 |
FLS2 |
8 |
3 |
1 |
2 |
1 |
5 |
ALDH |
7 |
3 |
2 |
3 |
2 |
3 |
Cathepsin B |
5 |
4 |
3 |
2 |
3 |
3 |
Cyclophilin |
4 |
5 |
3 |
2 |
4 |
1 |
Cytochrome C |
6 |
4 |
2 |
2 |
2 |
4 |
Drm-3 |
7 |
2 |
3 |
0 |
2 |
6 |
HCBT |
2 |
5 |
5 |
0 |
4 |
4 |
Plastocyanin |
4 |
6 |
2 |
2 |
2 |
4 |
Publications Supported by NSF Plant Genome Research Program Awards IOS-0077622, IOS-0605059, IOS-1025642 and IOS-1546625
1. Collmer, A., Lindeberg, M., Petnicki-Ocwieja, T., Schneider, D. J., and Alfano, J. R. 2002. Genomic mining type III secretion system effectors in Pseudomonas syringae yields new picks for all TTSS prospectors. Trends Microbiol. 10:462-469.
2. Fouts, D. E., Abramovitch, R. B., Alfano, J. R., Baldo, A. M., Buell, C. R., Cartinhour, S., Chatterjee, A. K., D'Ascenzo, M., Gwinn, M. L., Lazarowitz, S. G., Lin, N.-C., Martin, G. B., Rehm, A. H., Schneider, D. J., van Dijk, K., Tang, X., and Collmer, A. 2002. Genomewide identification of Pseudomonas syringae pv. tomato DC3000 promoters controlled by the HrpL alternative sigma factor. Proc. Natl. Acad. Sci. USA 99:2275-2280.
3. Jackson, R. W., Mansfield, J. W., Ammouneh, H., Dutton, L. C., Wharton, B., Ortiz-Barredo, A., Arnold, D. L., Tsiamis, G., Sesma, A., Butcher, D., Boch, J., Kim, Y. J., Martin, G. B., Tegli, S., Murillo, J., and Vivian, A. 2002. Location and activity of members of a family of virPphA homologues in pathovars of Pseudomonas syringae and P. savastanoi. Mol. Plant Pathol. 3:205-216.
4. Kim, Y.-J., Lin, N.-C., and Martin, G. B. 2002. Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109:589-598.
5. Petnicki-Ocwieja, T., Schneider, D. J., Tam, V. C., Chancey, S. T., Shan, L., Jamir, Y., Schechter, L. M., Buell, C. R., Tang, X., Collmer, A., and Alfano, J. R. 2002. Genomewide identification of proteins secreted by the Hrp type III protein secretion system of Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 99:7652-7657.
6. Abramovitch, R. B., Kim, Y. J., Chen, S., Dickman, M. B., and Martin, G. B. 2003. Pseudomonas type III effector AvrPtoB induces plant disease susceptibility by inhibition of host programmed cell death. EMBO J 22:60-69.
7. Alfano, J. R., and Guo, M. 2003. The Pseudomonas syringae Hrp (type III) protein secretion system: Advances in the new millenium. in: Plant-Microbe Interactions, Vol. 6. G. Stacey and N. T. Keen, eds. APS Press, St. Paul.
8. Badel, J. L., Nomura, K., Bandyopadhyay, S., Shimizu, R., Collmer, A., and He, S. Y. 2003. Pseudomonas syringae pv. tomato DC3000 HopPtoM (CEL ORF3) is important for lesion formation but not growth in tomato and is secreted and translocated by the Hrp type III secretion system in a chaperone-dependent manner. Mol. Microbiol. 49:1239-1251.
9. Buell, C. R., Joardar, V., Lindeberg, M., Selengut, J., Paulsen, I. T., Gwinn, M. L., Dodson, R. J., Deboy, R. T., Durkin, A. S., Kolonay, J. F., Madupu, R., Daugherty, S., Brinkac, L., Beanan, M. J., Haft, D. H., Nelson, W. C., Davidsen, T., Liu, J., Yuan, Q., Khouri, H., Fedorova, N., Tran, B., Russell, D., Berry, K., Utterback, T., Vanaken, S. E., Feldblyum, T. V., D'Ascenzo, M., Deng, W.-L., Ramos, A. R., Alfano, J. R., Cartinhour, S., Chatterjee, A. K., Delaney, T. P., Lazarowitz, S. G., Martin, G. B., Schneider, D. J., Tang, X., Bender, C. L., White, O., Fraser, C. M., and Collmer, A. 2003. The complete sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. USA 100:10181-10186.
10. Chatterjee, A., Cui, Y., Yang, H., Collmer, A., Alfano, J. R., and Chatterjee, A. K. 2003. GacA, the response regulator of a two-component system, acts as a master regulator in Pseudomonas syringae pv. tomato DC3000 by controlling regulatory RNA, transcriptional activators, and alternate sigma factors. Mol. Plant-Microbe Interact. 16:1106-1117.
11. Deng, W.-L., Rehm, A., Charkowski, A., Rojas, C. M., and Collmer, A. 2003. Pseudomonas syringae exchangeable effector loci: sequence diversity in representative pathovars and virulence function in P. syringae pv. syringae B728a. J. Bacteriol. 185:2592-2602.
12. Espinosa, A., Guo, M., Tam, V. C., Fu, Z. Q., and Alfano, J. R. 2003. The Pseudomonas syringae type III-secreted protein HopPtoD2 possesses protein tyrosine phosphatase activity and suppresses programmed cell death in plants. Mol. Microbiol. 49:377-387.
13. Alfano, J. R., and Collmer, A. 2004. Type III secretion system effector proteins: double agents in bacterial disease and plant defense. Annu. Rev. Phytopathol. 42:385-414.
14. D'Ascenzo, M. D., Collmer, A., and Martin, G. B. 2004. PeerGAD: A peer review-based and community-centric Web application for viewing and annotating genome sequences. Nucleic Acids Res. 32:3124-3135.
15. Espinosa, A., and Alfano, J. R. 2004. Disabling surveillance: bacterial type III secretion system effectors that suppress innate immunity. Cell Microbiol. 6:1027-1040.
16. He, P., Chintamanani, S., Chen, Z., Zhu, L., Kunkel, B. N., Alfano, J. R., Tang, X., and Zhou, J.-M. 2004. Activation of a COI1-dependent pathway in Arabidopsis by Pseudomonas syringae type III effectors and coronatine. Plant J. 37:589-602.
17.
Jamir, Y., Tang, X., and Alfano, J. R. 2004. The genome of Pseudomonas syringae pv. tomato DC3000 and functional genomic studies to better understand plant pathogenesis. in: Pseudomonas, Volume 1: Genomics, Life Style and Molecular Architecture. J. L. Ramos, eds. Kluwer/Plenum, London.
18. Jamir, Y., Guo, M., Oh, H.-S., Petnicki-Ocwieja, T., Chen, S., Tang, X., Dickman, M. B., Collmer, A., and Alfano, J. R. 2004. Identification of Pseudomonas syringae type III secreted effectors that suppress programmed cell death in plants and yeast. Plant J. 37:554-565.
19.
López-Solanilla, E., Bronstein, P. A., Schneider, A. R., and Collmer, A. 2004. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol. Microbiol. 54:353-365.
20. Schechter, L. M., Roberts, K. A., Jamir, Y., Alfano, J. R., and Collmer, A. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. J. Bacteriol. 186:543-555.
21. Shan, L., Oh, H. S., Chen, J., Guo, M., Zhou, J., Alfano, J. R., Collmer, A., Jia, X., and Tang, X. 2004. The HopPtoF locus of Pseudomonas syringae pv. tomato DC3000 encodes a type III chaperone and a cognate effector. Mol. Plant Microbe Interact. 17:447-455.
22. Wehling, M. D., Guo, M., Fu, Z. Q., and Alfano, J. R. 2004. The Pseudomonas syringae HopPtoV protein is secreted in culture and translocated into plant cells via the type III protein secretion system in a manner dependent on the ShcV type III chaperone. J. Bacteriol. 186:3621-3630.
23. Abramovitch, R. B., and Martin, G. B. 2005. AvrPtoB: a bacterial type III effector that both elicits and suppresses programmed cell death associated with plant immunity. FEMS Microbiol. Lett. 245:1-8.
24. Cohn, J. R., and Martin, G. B. 2005. Pseudomonas syringae pv. tomato type III effectors AvrPto and AvrPtoB promote ethylene-dependent cell death in tomato. Plant J. 44:139-154.
25. Joardar, V., Lindeberg, M., Schneider, D. J., Collmer, A., and Buell, C. R. 2005. Lineage specific regions in Pseudomonas syringae pv tomato DC3000. Mol. Plant Pathol. 6:53-64.
26. Joardar, V., Lindeberg, M., Jackson, R. W., Selengut, J., Dodson, R., Brinkac, L. M., Daugherty, S. C., DeBoy, R., Durkin, A. S., Giglio, M. G., Madupu, R., Nelson, W. C., Rosovitz, M. J., Sullivan, S., Haft, D. H., Creasy, T., Davidsen, T., Zafar, N., Zhou, L., Halpin, R., Holley, T., Khouri, H., Feldblyum, T., White, O., Fraser, C. M., Chatterjee, A. K., Cartinhour, S., Schneider, D. J., Mansfield, J., Collmer, A., and Buell, C. R. 2005. Whole genome sequence analysis of Pseudomonas syringae pv. phaseolicola 1448A reveals sequence divergence among pathovars in genes involved in virulence and mobile genetic elements. J. Bacteriol 187:6488-6498.
27. Lin, N. C., and Martin, G. B. 2005. An avrPto/avrPtoB mutant of Pseudomonas syringae pv. tomato DC3000 does not elicit Pto-specific resistance and is less virulent on tomato. Mol. Plant Microbe Interact. 18:43-51.
28. Lindeberg, M., Stavrinides, J., Chang, J. H., Alfano, J. R., Collmer, A., Dangl, J. L., Greenberg, J. T., Mansfield, J. W., and Guttman, D. S. 2005. Proposed guidelines for a unified nomenclature and phylogenetic analysis of type III Hop effector proteins in the plant pathogen Pseudomonas syringae. Mol. Plant-Microbe Interact. 18:275-282.
29. Oh, H.-S., and Collmer, A. 2005. Basal resistance against bacteria in Nicotiana benthamiana leaves is accompanied by reduced vascular staining and suppressed by multiple Pseudomonas syringae type III secretion system effector proteins. Plant J. 44:348-359.
30. Petnicki-Ocwiega, T., van Dijk, K., and Alfano, J. R. 2005. The hrpK operon of Pseudomonas syringae pv. tomato DC3000 encodes two proteins secreted by the type III (Hrp) protein secretion system: HopB1 and HrpK, a putative type III translocator. J. Bacteriol. 187:649-663.
31. Badel, J. L., Shimizu, R., Oh, H.-S., and Collmer, A. 2006. A Pseudomonas syringae pv. tomato avrE1/hopM1 mutant is severely reduced in growth and lesion formation in tomato. Mol. Plant Microbe Interact. 19:99-111.
32. Ferreira, A. O., Myers, C. R., Gordon, J. S., Martin, G. B., Vencato, M., Collmer, A., Wehling, M. D., Alfano, J. R., Moreno-Hagelsieb, G., Lamboy, W. F., DeClerck, G., Schneider, D. J., and Cartinhour, S. W. 2006. Whole-genome expression profiling defines the HrpL regulon of Pseudomonas syringae pv. tomato DC3000, allows de novo reconstruction of the Hrp cis element, and identifies novel co-regulated gene. Mol. Plant Microbe Interact. 19:1167-1179.
33. Lin, N. C., Abramovitch, R. B., Kim, Y. J., and Martin, G. B. 2006. Diverse AvrPtoB homologs from several Pseudomonas syringae pathovars elicit Pto-dependent resistance and have similar virulence activities. Appl. Environ. Microbiol. 72:702-712.
34. Lindeberg, M., Cartinhour, S., Myers, C. R., Schechter, L. M., Schneider, D. J., and Collmer, A. 2006. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol. Plant Microbe Interact. 19:1151-1158.
35. Robert-Seilaniantz, A., Shan, L. B., Zhou, J. M., and Tang, X. Y. 2006. The Pseudomonas syringae pv. tomato DC3000 type III effector HopF2 has a putative myristoylation site required for its avirulence and virulence functions. Mol. Plant-Microbe Interact. 19:130-138.
36. Sarkar, S. F., Gordon, J. S., Martin, G. B., and Guttman, D. S. 2006. Comparative genomics of host-specific virulence in Pseudomonas syringae. Genetics 174:1041-1056.
37. Schechter, L. M., Vencato, M., Jordan, K. L., Schneider, S. E., Schneider, D. J., and Collmer, A. 2006. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol. Plant-Microbe Interact. 19:1180-1192.
38. Vencato, M., Tian, T., Alfano, J. R., Buell, C. R., Cartinhour, S., DeClerck, G. A., Guttman, D. S., Stavrinides, J., Joardar, V., Lindeberg, M., Bronstein, P. A., Mansfield, J. W., Myers, C. R., Collmer, A., and Schneider, D. J. 2006. Bioinformatics-enabled identification of the HrpL regulon and type III secretion system effector proteins of Pseudomonas syringae pv. phaseolicola 1448A. Mol. Plant Microbe Interact. 19:1193-1206.
39. Fu, Z. Q., Guo, M., Jeong, B. R., Tian, F., Elthon, T. E., Cerny, R. L., Staiger, D., and Alfano, J. R. 2007. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447:284-8.
40. Lin, N.-C., and Martin, G. B. 2007. Pto/Prf-mediated recognition of AvrPto and AvrPtoB restricts the ability of diverse Pseudomonas syringae pathovars to infect tomato. Mol. Plant Microbe Interact. 20:806-815.
41. Rosebrock, T. R., Zeng, L., Brady, J. J., Abramovitch, R. B., Xiao, F., and Martin, G. B. 2007. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448:370-374.
42. Wei, C.-F., Kvitko, B. H., Shimizu, R., Crabill, E., Alfano, J. R., Lin, N.-C., Martin, G. B., Huang, H.-C., and Collmer, A. 2007. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 51:32-46.
43. Xiao, F., He, P., Abramovitch, R. B., Dawson, J. E., Nicholson, L. K., Sheen, J., and Martin, G. B. 2007. The N-terminal region of Pseudomonas type III effector AvrPtoB elicits Pto-dependent immunity and has two distinct virulence determinants. Plant J. 52:595-614.
44. Collmer, C. W., Lindeberg, M., and Collmer, A. 2008. Gene Ontology (GO) for microbe-host interactions and its use in ongoing annotation of three Pseudomonas syringae genomes via the Pseudomonas-Plant Interaction (PPI) web site. in: Pseudomonas syringae Pathovars and Related Pathogens - Identification, Epidemiology and Genomics M. Fatmi, A. Collmer, N. S. Iacobellis, J. Mansfield, J. Murillo, N. W. Schaad and M. Ullrich, eds. Springer.
45. Collmer, A., Kvitko, B. H., Morello, J. E., Munkvold, K. R., Oh, H.-S., and Wei, C.-F. 2008. Exploring the functions of proteins secreted by the Hrp type III secretion system of Pseudomonas syringae. in: Pseudomonas syringae Pathovars and Related Pathogens - Identification, Epidemiology and Genomics. M. Fatmi, A. Collmer, N. S. Iacobellis, J. Mansfield, J. Murillo, N. W. Schaad and M. Ullrich, eds. Springer.
46. Lindeberg, M., Schneider, D. J., Cartinhour, S., and Collmer, A. 2008. Genomic analysis of Pseudomonas syringae pathovars: identification of virulence genes and associated regulatory elements using pattern-based searches and genome comparison. in: Pseudomonas syringae Pathovars and Related Pathogens - Identification, Epidemiology and Genomics. M. Fatmi, A. Collmer, N. S. Iacobellis, J. Mansfield, J. Murillo, N. W. Schaad and M. Ullrich, eds. Springer.
47. Lindeberg, M., Myers, C. R., Collmer, A., and Schneider, D. J. 2008. Roadmap to new virulence determinants in Pseudomonas syringae: Insights from comparative genomics and genome organization. Mol. Plant-Microbe Interact. 21:685-700.
48. Munkvold, K. R., Martin, M. E., Bronstein, P. A., and Collmer, A. 2008. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol. Plant-Microbe Interact. 21:490-502.
49. Shan, L., He, P., Li, J., Heese, A., Peck, S. C., Nurnberger, T., Martin, G. B., and Sheen, J. 2008. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4:17-27.
50. Almeida, N. F., Yan, S., Lindeberg, M., Studholme, D. J., Schneider, D. J., Condon, B., Liu, H., Viana, C. J., Warren, A., Evans, C., Kemen, E., MacLean, D., Angot, A., Martin, G. B., Jones, J. D., Collmer, A., Setubal, J. C., and Vinatzer, B. A. 2009. A draft genome sequence of Pseudomonas syringae pv. tomato strain T1 reveals a repertoire of type III related genes significantly divergent from that of Pseudomonas syringae pv. tomato strain DC3000. Mol. Plant-Microbe Interact. 22:52-62.
51. Chakravarthy, S., Velasquez, A. C., and Martin, G. B. 2009. Assay for pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) in plants. J. Vis. Exp. 31:http://www.jove.com/index/Details.stp?ID=1442, doi: 10.3791/1442.
52. Collmer, A., Schneider, D. J., and Lindeberg, M. 2009. Lifestyles of the effector rich: genome-enabled characterization of bacterial plant pathogens. Plant Physiol 150:1623-30.
53. Cunnac, S., Lindeberg, M., and Collmer, A. 2009. Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12:53-60.
54. Dong, J., Xiao, F., Fan, F., Gu, L., Cang, H., Martin, G. B., and Chai, J. 2009. Crystal structure of the complex between Pseudomonas effector AvrPtoB and the tomato Pto kinase reveals both a shared and a unique interface compared with AvrPto-Pto. Plant Cell 21:1846-59.
55. Kim, J.-G., Roden, J. A., Taylor, K. W., Aarkre, C. D., Su, B., Lalonde, S., Kirik, A., Chen, Y., Baranage, G., McLane, H., Martin, G. B., and Mudgett, M. B. 2009. Xanthomonas T3S effector XopN suppresses PAMP-triggered immunity and interacts with a tomato atypical receptor-like kinase and TFT1. Plant Cell 21:1305-23.
56. Kvitko, B. H., and Collmer, A. 2009. Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. in: Methods in Molecular Biology: Plant Immunity. J. McDowell, eds. Humana Press.
57. Kvitko, B. H., Park, D. H., Velásquez, A. C., Wei, C.-F., Russell, A. B., Martin, G. B., Schneider, D. J., and Collmer, A. 2009. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathogens 5:e1000388.
58. Lindeberg, M., Cunnac, S., and Collmer, A. 2009. The evolution of Pseudomonas syringae host specificity and type III effector repertoires. Mol. Plant Pathol. 10:767-775.
59. Munkvold, K. R., and Martin, G. B. 2009. Advances in experimental methods for elucidating Pseudomonas syringae effector function with a focus on AvrPtoB. Mol. Plant Pathol. 10:777-793.
60. Munkvold, K. R., Russell, A. B., Kvitko, B. H., and Collmer, A. 2009. Pseudomonas syringae pv. tomato DC3000 type III effector HopAA1-1 functions redundantly with chlorosis-promoting factor PSPTO4723 to produce bacterial speck lesions in host tomato. Mol. Plant Microbe Interact. 22:1341–1355.
61. Velasquez, A. C., Chakravarthy, S., and Martin, G. B. 2009. Virus-induced gene silencing (VIGS) in Nicotiana benthamiana and tomato. J. Vis. Exp. 28:http://www.jove.com/index/details.stp?id=1292, doi: 10.3791/1292.
62. Chakravarthy, S., Velásquez, A. C., Ekengren, S. K., Collmer, A., and Martin, G. B. 2010. Identification of Nicotiana benthamiana genes involved in PAMP-triggered immunity. Mol. Plant-Microbe Interact.:715-726.
63. Nguyen, N. P., Chakravarthy, S., Velásquez, A. C., McLane, H. S., Zeng, L., Nakayashiki, H., Park, D. H., Collmer, A., and Martin, G. B. 2010. Methods to study PAMP-triggered immunity using tomato and Nicotiana benthamiana. Mol. Plant Microbe Interact. 23:991-999.
64. Oh, H.-S., Park, D. H., and Collmer, A. 2010. Components of the Pseudomonas syringae type III secretion system can suppress and may elicit plant innate immunity Mol. Plant-Microbe Interact. 23:727-739.
65. Schneider, D. J., and Collmer, A. 2010. Studying plant-pathogen interactions in the genomics era: beyond molecular Koch's postulates to systems biology. Annu. Rev. Phytopathol. 48:457-479.
66. Kvitko, B. H., and Collmer, A. 2011. Construction of Pseudomonas syringae pv. tomato DC3000 mutant and polymutant strains. Pages 109-128 in: Methods in Molecular Biology: Plant Immunity. J. M. McDowell, eds. Springer Science+Business Media
67. Park, D. H., Mirabella, R., Bronstein, P. A., Preston, G. M., Haring, M. A., Lim, C. K., Collmer, A., and Schuurink, R. C. 2010. Mutations in gamma-aminobutyric acid (GABA) transaminase genes in plants or Pseudomonas syringae reduce bacterial virulence. Plant J. 64:318–330
68. Kelley, B. S., Lee, S. J., Damasceno, C. M., Chakravarthy, S., Kim, B. D., Martin, G. B., and Rose, J. K. 2010. A secreted effector protein (SNE1) from Phytophthora infestans is a broadly acting suppressor of programmed cell death. Plant J 62:357-66.
69. Cunnac, S., Chakravarthy, S., Kvitko, B. H., Russell, A. B., Martin, G. B., and Collmer, A. 2011. Genetic disassembly and combinatorial reassembly identify a minimal functional repertoire of type III effectors in Pseudomonas syringae. Proc. Natl. Acad. Sci. U S A 108:2975-80.
70. Cheng, W., Munkvold, K. R., Gao, H., Mathieu, J., Schwizer, S., Wang, S., Yan, Y. B., Wang, J., Martin, G. B., and Chai, J. 2011. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe 10:616-26
71. Zeng, L., Velasquez, A. C., Munkvold, K. R., Zhang, J., and Martin, G. B. 2011. A tomato LysM receptor-like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J 69:92-103
72. Lindeberg M, Cunnac S, Collmer A. 2012. Pseudomonas syringae type III effector repertoires: last words in endless arguments. Trends Microbiol. 20:199-208.
73. Lindeberg M. 2012. Genome-enabled perspectives on the composition, evolution, and expression of virulence determinants in bacterial plant pathogens.. Ann Rev Phytopath. 50:111-132.
74. Schechter LM, Valenta JC, Schneider DJ, Collmer A, Sakk E. 2012. Functional and Computational Analysis of Amino Acid Patterns Predictive of Type III Secretion System Substrates in Pseudomonas syringae. PLoS One 7.
75. Wei HL, Collmer A. 2012. Multiple lessons from the multiple functions of a regulator of type III secretion system assembly in the plant pathogen Pseudomonas syringae. Mol Microbiol.
76. Bao, Z., Stodghill, P.V., Myers, C.R., Lam, H., Wei, H.-L., Chakravarthy, S., Kvitko, B.H., Collmer, A., Cartinhour, S.W., Schweitzer, P., and Swingle, B. 2014. Genomic Plasticity Enables Phenotypic Variation of Pseudomonas syringae pv. tomato DC3000. PLOS ONE 9:e86628.
77. Velásquez, A. C., and Martin, G. B. 2012. Molecular mechanisms involved in the interaction between tomato and Pseudomonas syringae pv. tomato. in: Molecular Plant Immunity. G. Sessa, eds. Wiley-Blackwell.
78. Martin, G. B. 2012. Suppression and activation of the plant immune system by Pseudomonas syringae effectors AvrPto and AvrPtoB. in: Effectors in Plant-Microbe Interactions. F. Martin and S. Kamoun, eds. Wiley-Blackwell
79. Rodriguez-Herva, J. J., Gonzalez-Melendi, P., Cuartas-Lanza, R., Antunez-Lamas, M., Rio-Alvarez, I., Li, Z., Lopez-Torrejon, G., Diaz, I., Del Pozo, J. C., Chakravarthy, S., Collmer, A., Rodriguez-Palenzuela, P., and Lopez-Solanilla, E. 2012. A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell Microbiol 14:669-681.
80. Bombarely, A., Rosli, H. G., Vrebalov, J., Moffett, P., Mueller, L. A., and Martin, G. B. 2012. A draft genome sequence of Nicotiana benthamiana to enhance molecular plant-microbe biology research. Mol. Plant Microbe Interact 12:1523-30.
81. de la Torre, F., Gutierrez-Beltran, E., Pareja-Jaime, Y., Chakravarthy, S., Martin, G. B., and Del Pozo, O. 2013. The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell 25:2748-2764.
82. Singh, D. K., Calvino, M., Brauer, E. K., Fernandez-Pozo, N., Strickler, S., Yalamanchili, R. D., Suzuki, H., Aoki, K., Shibata, D., Stratmann, J. W., Popescu, G. V., Mueller, L., and Popescu, S. C. 2013. The tomato kinome and the TOKN ORFeome: novel resources for the study of kinases and signal transduction in tomato and Solanaceae. Mol Plant Microbe Interact.
83. Wei, H.-L., Chakravarthy, S., Worley, J. N., and Collmer, A. 2013. Consequences of flagellin export through the type III secretion system of Pseudomonas syringae reveal a major difference in the innate immune systems of mammals and the model plant Nicotiana benthamiana. Cell. Microbiol. 15:601-618.
84. Worley, J. N., Russell, A. B., Wexler, A. G., Bronstein, P. A., Kvitko, B. H., Krasnoff, S. B., Munkvold, K. R., Swingle, B., Gibson, D. M., and Collmer, A. 2013. Pseudomonas syringae pv. tomato DC3000 CmaL (PSPTO4723), a DUF1330 family member, is needed to produce L-allo-isoleucine, a precursor for the phytotoxin coronatine. J Bacteriol. 195:287-296.
85. Saha, S., and Lindeberg, M. 2013. Bound to Succeed: Transcription Factor Binding-site Prediction and Its Contribution to Understanding Virulence and Environmental Adaptation in Bacterial Plant Pathogens. Mol Plant Microbe Interact 26:1123-1130.
86. Gardiner, D.M., Stiller, J., Covarelli, L., Lindeberg, M., Shivas, R.G., and Manners, J.M. 2013. Genome Sequences of Pseudomonas spp. Isolated from Cereal Crops. Genome Announcements 1(3).
87. Rosli HG, Zheng Y, Pombo MA, Zhong S, Bombarely A, Fei Z, Collmer A, Martin
GB. Transcriptomics-based screen for genes induced by flagellin and repressed by
pathogen effectors identifies a cell wall-associated kinase involved in plant
immunity. Genome Biol. 2013 Dec 20;14(12):R139.
88. Clarke CR, D. Chinchilla, S. R. Hind, F. Taguchi, R. Miki, Y. Ichinose, G. B. Martin, S. Leman, G. Felix and B. A. Vinatzer (2013). Allelic variation in two distinct Pseudomonas syringae flagellin epitopes modulates the strength of plant immune responses but not bacterial motility. New Phytologist 200:847-860
89. Veluchamy, S., Hind, S.R., Dunham, D.M., Martin, G.B., and Panthee, D.R. 2014. Natural variation for responsiveness to flg22, flgII-28, and csp22 and Pseudomonas syringae pv. tomato in heirloom tomatoes. PloS one 9:e106119.
90. Pombo, M.A., Zheng, Y., Fernandez-Pozo, N., Dunham, D.M., Fei, Z., and Martin, G.B. 2014. Transcriptomic analysis reveals tomato genes whose expression is induced specifically during effector-triggered immunity and identifies the Epk1 protein kinase which is required for the host response to three bacterial effector proteins. Genome biology 15:492
91. Mathieu, J., Schwizer, S., and Martin, G.B. 2014. Pto kinase binds two domains of AvrPtoB and its proximity to the effector E3 ligase determines if it evades degradation and activates plant immunity. PLoS pathogens 10:e1004227.
92. Jones, L.A., Saha, S., Collmer, A., Smart, C.D., and Lindeberg, M. 2014. Genome-Assisted Development of a Diagnostic Protocol for Distinguishing High Virulence Pseudomonas syringae pv. tomato Strains. Plant Disease 99:527-534.
93. Rosli, H.G., and Martin, G.B. 2015. Functional genomics of tomato for the study of plant immunity. Briefings in Functional Genomics 14:291-301
Fernandez-Pozo, N., H.G. Rosli, G.B. Martin, and L.A. Mueller, 2015. The SGN VIGS Tool: User-friendly software to design virus-induced gene silencing (VIGS) constructs for functional genomics. Mol Plant, 8:486-488.
94. Wei, H.-L., Chakravarthy, S., Mathieu, J., Helmann, Tyler C., Stodghill, P., Swingle, B., Martin, Gregory B., and Collmer, A. 2015. Pseudomonas syringae pv. tomato DC3000 Type III Secretion Effector Polymutants Reveal an Interplay between HopAD1 and AvrPtoB. Cell Host & Microbe 17:752-762.
95. Worley, J.N., Pombo, M.A., Zheng, Y., Dunham, D.M., Myers, C.R., Fei, Z., and Martin, G.B. 2016. A novel method of transcriptome interpretation reveals a quantitative suppressive effect on tomato immune signaling by two domains in a single pathogen effector protein. BMC Genomics 17:229.
96. Kraus, Christine M., Munkvold, Kathy R., and Martin, Gregory B. 2016. Natural Variation in Tomato Reveals Differences in the Recognition of AvrPto and AvrPtoB Effectors from Pseudomonas syringae. Molecular plant 9:639-649.
96. Hind, S.R., S.R. Strickler, P.C. Boyle, D.M. Dunham, Z. Bao, I.M. O'Doherty, J.A. Baccile, J.S. Hoki, E.G. Viox, C.R. Clarke, B.A. Vinatzer, F.C. Schroeder, and G.B. Martin Tomato receptor FLAGELLIN-SENSING 3 binds flgII-28 and activates the plant immune system. Nat Plants, 2016. 2:16128
97. Boyle, P.C., S. Schwizer, S.R. Hind, C.M. Kraus, S. De la Torre Diaz, B. He, and G.B. Martin, 2016. Detecting N-myristoylation and S-acylation of host and pathogen proteins in plants using click chemistry. Plant Methods, 12:38.
98. Pombo, M.A., Zheng, Y., Fei, Z., Martin, G.B., and Rosli, H.G. 2017. Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem 7:44905.
99. Schwizer, S., Kraus, C.M., Dunham, D.M., Zheng, Y., Fernandez-Pozo, N., Pombo, M.A., Fei, Z., Chakravarthy, S., and Martin, G.B. 2017. The Tomato Kinase Pti1 Contributes to Production of Reactive Oxygen Species in Response to Two Flagellin-Derived Peptides and Promotes Resistance to Pseudomonas syringae Infection. Molecular Plant-Microbe Interactions 30:725-738.
100. Wei, H.-L., and Collmer, A. 2017. Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors. Molecular Plant Pathology 19:1779-1794
101. Kraus, C.M., Mazo-Molina, C., Smart, C.D., and Martin, G.B. 2017. Pseudomonas syringae pv. tomato Strains from New York Exhibit Virulence Attributes Intermediate Between Typical Race 0 and Race 1 Strains. Plant Disease 101:1442-1448.
102. Jacobs, T.B., Zhang, N., Patel, D., and Martin, G.B. 2017. Generation of a Collection of Mutant Tomato Lines Using Pooled CRISPR Libraries. Plant Physiology 174:2023-2037
103. Hatsugai, N., D. Igarashi, K. Mase, Y. Lu, Y. Tsuda, S. Chakravarthy, H.L. Wei, J.W. Foley, A. Collmer, J. Glazebrook, and F. Katagiri, 2017. A plant effector-triggered immunity signaling sector is inhibited by pattern-triggered immunity. EMBO J, 36:2758-2769
104. Bhattarai, K., F.J. Louws, J.D. Williamson, and D.R. Panthee, 2017. Resistance to Xanthomonas perforans race T4 causing bacterial spot in tomato breeding lines. Plant Pathol, 66:1103-1109
105. Hwang, I.S., J.J. Brady, G.B. Martin, and C.-S. Oh, 2017. Ser360 and Ser364 in the kinase domain of tomato SlMAPKKKα are critical for programmed cell death associated with plant immunity. Plant Path. J., 33:163-169
106. Hind, S.R., Hoki, J.S., Baccile, J.S., Boyle, P., Schroeder, F.C., and Martin, G.B. 2017. Detecting the Interaction of Peptide Ligands with Plant Membrane Receptors. Current Protocols in Plant Biology 2:240-269
107. Pombo, M.A., Y. Zheng, Z. Fei, G.B. Martin, and H.G. Rosli, 2017. Use of RNA-seq data to identify and validate RT-qPCR reference genes for studying the tomato-Pseudomonas pathosystem. Sci Rep, 7:44905.
108. Chakravarthy, S., Worley, J.N., Montes-Rodriguez, A., and Collmer, A. 2018. Pseudomonas syringae pv. tomato DC3000 polymutants deploying coronatine and two type III effectors produce quantifiable chlorotic spots from individual bacterial colonies in Nicotiana benthamiana leaves. Molecular Plant Pathology:19:935-947.
109. Wei, H.-L., Zhang, W., and Collmer, A. 2018. Modular Study of the Type III Effector Repertoire in Pseudomonas syringae pv. tomato DC3000 Reveals a Matrix of Effector Interplay in Pathogenesis. Cell Reports 23:1630-1638
110. Adhikari, P., 2018, Mapping QTLs derived from Solanum pimpinellifolium LA3707 for bacterial spot disease resistance and fruit morphology in tomato. North Carolina State University.
111. Gonda, I., H. Ashrafi, D.A. Lyon, S.R. Strickler, A.M. Hulse-Kemp, Q. Ma, H. Sun, K. Stoffel, A.F. Powell, S. Futrell, T.W. Thannhauser, Z. Fei, A.E. Van Deynze, L.A. Mueller, J.J. Giovannoni, and M.R. Foolad. 2019. Sequencing-based bin map construction of a tomato mapping population, facilitating high-resolution quantitative trait loci detection. Plant Genome, 12.
112. Zheng, Y., N. Zhang, G.B. Martin, and Z. Fei, 2019. Plant Genome Editing Database (PGED): A call for submission of information about genome-edited plant mutants. Mol Plant, 12:127-129
113. Adhikari, P., T.B. Adhikari, S. Timilsina, I. Meadows, J.B. Jones, D.R. Panthee, and F.J. Louws, 2019. Phenotypic and genetic diversity of Xanthomonas perforans populations from tomato in North Carolina. Phytopathol, 109:1533-1543.
114. Eckshtain-Levi, N., M. Lindeberg, G.E. Vallad, and G.B. Martin, 2019. The tomato Pto gene confers resistance to Pseudomonas floridensis, an emergent plant pathogen with just nine type III effectors. Plant Pathology, 68:977-984
115. Mazo-Molina, C., S. Mainiero, S.R. Hind, C.M. Kraus, M. Vachev, F. Maviane-Macia, M. Lindeberg, S. Saha, S.R. Strickler, A. Feder, J.J. Giovannoni, C.D. Smart, N. Peeters, and G.B. Martin, 2019. The Ptr1 locus of Solanum lycopersicoides confers resistance to race 1 strains of Pseudomonas syringae pv. tomato and to Ralstonia pseudosolanacearum by recognizing the type III effectors AvrRpt2 and RipBN. Mol Plant Microbe Interact, 32:949-960
116. Roberts, R., S. Mainiero, A.F. Powell, A.E. Liu, K. Shi, S.R. Hind, S.R. Strickler, A. Collmer, and G.B. Martin, 2019. Natural variation for unusual host responses and flagellin-mediated immunity against Pseudomonas syringae in genetically diverse tomato accessions. New Phytol, 223:447-461
117. Roberts, R., S.R. Hind, K.F. Pedley, B.A. Diner, M.J. Szarzanowicz, D. Luciano-Rosario, B.B. Majhi, G. Popov, G. Sessa, C.S. Oh, and G. Martin, 2019. Mai1 protein acts between host recognition of pathogen effectors and mitogen-activated protein kinase signaling. Mol Plant Microbe Interact, 32:1496-1507
118. Giska, F. and G.B. Martin, 2019. PP2C phosphatase Pic1 negatively regulates the phosphorylation status of Pti1b kinase, a regulator of flagellin-triggered immunity in tomato. Biochem J, 476:1621-1635.
119. Wei, H.-L., Zhang, W., and Collmer, A. 2018. Modular Study of the Type III Effector Repertoire in Pseudomonas syringae pv. tomato DC3000 Reveals a Matrix of Effector Interplay in Pathogenesis. Cell Reports 23:1630-1638
120. Zhang , N., H.M. Roberts, J. Van Eck, and G.B. Martin, 2020. Generation and molecular characterization of CRISPR/Cas9-induced mutations in 63 immunity-associated genes in tomato reveals specificity and a range of gene modifications. Front. Plant Sci., 11:1-13
121. Zhang, J. and D.R. Panthee, 2020. PyBSASeq: a simple and effective algorithm for bulked segregant analysis with whole-genome sequencing data. BMC Bioinformatics, 21:99
122. Adhikari, P., T.B. Adhikari, F.J. Louws, and D.R. Panthee, 2020. Advances and challenges in bacterial spot resistance breeding in tomato (Solanum lycopersicum L.). Int J Mol Sci, 21.
123. Zhang, N., M.A. Pombo, H.G. Rosli, and G.B. Martin, 2020. Tomato wall-associated kinase SlWak1 acts in an Fls2- and Fls3-dependent manner to promote apoplastic immune responses to Pseudomonas syringae. bioRxiv, 10.1101/2020.01.27.921460:2020.01.27.921460
124. Powell, A.F., L.E. Courtney, M.H.-W. Schmidt, A. Feder, A. Vogel, Y. Xu, D.A. Lyon, K.E. Dumschott, M. McHale, R. Suplice, K. Bao, A. Duhan, A. Hallab, A.K. Denton, L.A. Mueller, S. Alseekh, J. Lie, C. Martin, A.R. Fernie, S.R. Hind, G.B. Martin, Z. Fei, J.J. Giovannoni, S.R. Strickler, and B. Usadel, 2020. A Solanum lycopersicoides reference genome facilitates biological discovery in tomato. bioRxiv,:2020.04.16.039636.
125. Roberts, R., A.E. Liu, L. Wan, A.M. Geiger, S.R. Hind, H.G. Rosli, and G.B. Martin, Molecular characterization of differences between the tomato immune receptors Fls3 and Fls2. 2020. bioRxiv 10.1101/2020.02.04.934133:2020.02.04.934133.
126. Mazo-Molina, C., S.M. S., B.J. Benjamin J. Haefner, R. Bednarek, R. Zhang, A. Feder, K. Shi, S.R. Strickler, and G.B. Martin, 2020. Ptr1 evolved convergently with RPS2 and Mr5 to mediate recognition of AvrRpt2 in diverse solanaceous species. bioRxiv, https://www.biorxiv.org/content/10.1101/2020.03.05.979484v1.
|
